-Search query
-Search result
Showing 1 - 50 of 426 items for (author: zhang & mm)
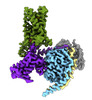
EMDB-43647: 
CryoEM structure of Gi-coupled TAS2R14 with cholesterol and an intracellular tastant
Method: single particle / : Kim Y, Gumpper RH, Roth BL
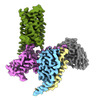
EMDB-43650: 
CryoEM structure of Ggust-coupled TAS2R14 with cholesterol and an intracellular tastant
Method: single particle / : Kim Y, Gumpper RH, Roth BL

EMDB-43656: 
CryoEM structure of Gi-coupled TAS2R14 with cholesterol and an intracellular tastant (Locally refined map)
Method: single particle / : Kim Y, Gumpper RH, Roth BL

EMDB-43657: 
CryoEM structure of Ggust-coupled TAS2R14 with cholesterol and an intracellular tastant (Locally refined map)
Method: single particle / : Kim Y, Gumpper RH, Roth BL

PDB-8vy7: 
CryoEM structure of Gi-coupled TAS2R14 with cholesterol and an intracellular tastant
Method: single particle / : Kim Y, Gumpper RH, Roth BL

PDB-8vy9: 
CryoEM structure of Ggust-coupled TAS2R14 with cholesterol and an intracellular tastant
Method: single particle / : Kim Y, Gumpper RH, Roth BL

EMDB-17630: 
ABCB1 L335C mutant (mABCB1) in the inward facing state bound to AAC
Method: single particle / : Parey K, Januliene D, Gewering T, Moeller A, Vecchis D, Striednig B, Hilbi H, Schaefer LV, Kuprov I, Bordignon E, Seeger MA

PDB-8pee: 
ABCB1 L335C mutant (mABCB1) in the inward facing state bound to AAC
Method: single particle / : Parey K, Januliene D, Gewering T, Moeller A
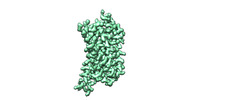
EMDB-43732: 
momSalB bound Kappa Opioid Receptor in complex Gi1
Method: single particle / : Fay JF, Che T

EMDB-43733: 
GR89,696 bound Kappa Opioid Receptor in complex with Gz
Method: single particle / : Fay JF, Che T

EMDB-43734: 
GR89,696 bound Kappa Opioid Receptor in complex with gustducin
Method: single particle / : Fay JF, Che T
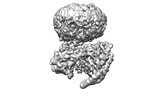
EMDB-42023: 
GPR3 Orphan G-coupled Protein Receptor in complex with Dominant Negative Gs.
Method: single particle / : Russell IC, Belousoff MJ, Sexton P

EMDB-41830: 
Lipidated recombinant apolipoprotein E4
Method: single particle / : Strickland MR, Rau M, Summers B, Basore K, Wulf II J, Jiang H, Chen Y, Ulrich JD, Randolph GJ, Zhang R, Fitzpatrick JAJ, Cashikar AG, Holtzman DM

EMDB-41831: 
Gradient-fixed lipidated recombinant apolipoprotein E4
Method: single particle / : Strickland MR, Rau M, Summers B, Basore K, Wulf II J, Jiang H, Chen Y, Ulrich JD, Randolph GJ, Zhang R, Fitzpatrick JAJ, Cashikar AG, Holtzman DM

EMDB-41048: 
Lassa GPC Trimer in complex with Fab 8.11G and nanobody D5
Method: single particle / : Gorman J, Kwong PD

PDB-8t5c: 
Lassa GPC Trimer in complex with Fab 8.11G and nanobody D5
Method: single particle / : Gorman J, Kwong PD
EMDB-41617: 
CryoEM structure of PI3Kalpha
Method: single particle / : Valverde R, Shi H, Holliday M, Sun M

EMDB-41144: 
Cryo-EM Structure of GPR61-G protein complex stabilized by scFv16
Method: single particle / : Lees JA, Dias JM, Han S

EMDB-41145: 
Cryo-EM Structure of GPR61-
Method: single particle / : Lees JA, Dias JM, Han S

PDB-8tb0: 
Cryo-EM Structure of GPR61-G protein complex stabilized by scFv16
Method: single particle / : Lees JA, Dias JM, Han S

EMDB-41876: 
momSalB bound Kappa Opioid Receptor in complex with GoA
Method: single particle / : Fay JF, Che T

EMDB-28115: 
Western Equine Encephalitis Virus-Like Particle in Complex with SKW19 Fab
Method: single particle / : Pletnev S, Tsybovsky Y, Verardi R, Roedeger M, Kwong P

EMDB-28116: 
Western Equine Encephalitis Virus-Like Particle in Complex with SKW24 Fab
Method: single particle / : Pletnev S, Tsybovsky Y, Verardi R, Roedeger M, Kwong PD

EMDB-28117: 
Eastern Equine Encephalitis Virus-Like Particle in Complex with SKE26 Fab
Method: single particle / : Pletnev S, Verardi R, Roedeger M, Kwong P

EMDB-34393: 
Cryo-EM structure of Abeta2 fibril polymorph2
Method: helical / : Xia WC, Zhang MM, Liu C

EMDB-34392: 
Cryo-EM structure of Abeta2 fibril polymorph1
Method: helical / : Xia WC, Zhang MM, Liu C

EMDB-15687: 
The ABCB1 L335C mutant (mABCB1) in the Apo state
Method: single particle / : Parey K, Januliene D, Gewering T, Moeller A

PDB-8avy: 
The ABCB1 L335C mutant (mABCB1) in the Apo state
Method: single particle / : Parey K, Januliene D, Gewering T, Moeller A
EMDB-28802: 
Subtomographic average of flagellar axoneme doublet from Trypanosoma brucei FAP106 knockdown mutant
Method: subtomogram averaging / : Shimogawa MM, Wang H
EMDB-28803: 
Subtomographic average of flagellar axoneme doublet from Trypanosoma brucei Wildtype
Method: subtomogram averaging / : Shimogawa MM, Wang H

EMDB-28804: 
Subtomographic average of flagellar axoneme doublet from Trypanosoma brucei MC8 Knockdown mutant
Method: subtomogram averaging / : Shimogawa MM, Wang H
EMDB-28805: 
Subtomographic average of flagellar axoneme doublet from Trypanosoma brucei Wildtype Control 2
Method: subtomogram averaging / : Shimogawa MM, Wang H

EMDB-41302: 
Lassa GPC trimer in complex with Fab GP23
Method: single particle / : Gorman J, Kwong PD
EMDB-28118: 
Cryo-EM structure of Antibody SKW11 in complex with Western Equine Encephalitis Virus-Like Particle
Method: single particle / : Cerutti G, Verardi R, Roederer M, Shapiro L
EMDB-28119: 
Cryo-EM structure of Antibody SKT05 in complex with Western Equine Encephalitis Virus-like Particle
Method: single particle / : Cerutti G, Verardi R, Roederer M, Shapiro L

EMDB-28187: 
Cryo-EM I1 reconstruction of antibody SKV09 in complex with VEEV alphavirus VLP
Method: single particle / : Casner RG, Verardi R, Roederer M, Shapiro L

EMDB-28188: 
Cryo-EM I1 reconstruction of antibody SKV16 in complex with VEEV alphavirus VLP
Method: single particle / : Casner RG, Verardi R, Roederer M, Shapiro L

EMDB-27757: 
Cryo-EM Structure of Eastern Equine Encephalitis Virus in complex with SKE26 Fab
Method: single particle / : Pletnev S, Verardi R, Roedeger M, Kwong P

EMDB-28056: 
Venezuelan equine encephalitis virus-like particle in complex with Fab SKT05
Method: single particle / : Tsybovsky Y, Pletnev S, Verardi R, Roederer M, Kwong PD

EMDB-28058: 
Venezuelan equine encephalitis virus-like particle in complex with Fab SKT05, local refinement
Method: single particle / : Tsybovsky Y, Pletnev S, Verardi R, Roederer M, Kwong PD

EMDB-28059: 
Venezuelan equine encephalitis virus-like particle in complex with Fab SKT20
Method: single particle / : Tsybovsky Y, Pletnev S, Verardi R, Roederer M, Kwong PD

EMDB-28060: 
Venezuelan equine encephalitis virus-like particle in complex with Fab SKT-20, local refinement
Method: single particle / : Tsybovsky Y, Pletnev S, Verardi R, Roederer M, Kwong PD

PDB-8dwo: 
Cryo-EM Structure of Eastern Equine Encephalitis Virus in complex with SKE26 Fab
Method: single particle / : Pletnev S, Verardi R, Roedeger M, Kwong P

PDB-8eeu: 
Venezuelan equine encephalitis virus-like particle in complex with Fab SKT05
Method: single particle / : Tsybovsky Y, Pletnev S, Verardi R, Roederer M, Kwong PD

PDB-8eev: 
Venezuelan equine encephalitis virus-like particle in complex with Fab SKT-20
Method: single particle / : Tsybovsky Y, Pletnev S, Verardi R, Roederer M, Kwong PD

EMDB-27389: 
Cryo-EM Structure of Western Equine Encephalitis Virus
Method: single particle / : Pletnev S, Verardi R, Roedeger M, Kwong P

EMDB-27390: 
Cryo-EM Structure of Western Equine Encephalitis Virus VLP in complex with SKW19 fab
Method: single particle / : Pletnev S, Tsybovsky Y, Verardi R, Roedeger M, Kwong P

EMDB-27391: 
Asymmetric Unit of Western Equine Encephalitis Virus
Method: single particle / : Pletnev S, Verardi R, Roedeger M, Kwong P

EMDB-27392: 
Cryo-EM Structure of Western Equine Encephalitis Virus VLP in complex with SKW24 fab
Method: single particle / : Pletnev S, Tsybovsky Y, Verardi R, Roedeger M, Kwong PD
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
